产品号 #200-0510_C
一次性使用、免维持的人诱导多能干细胞,冷冻

cGMP, stabilized feeder-free maintenance medium for human ES and iPS cells
Compatible antibodies for purity assessment of isolated cells
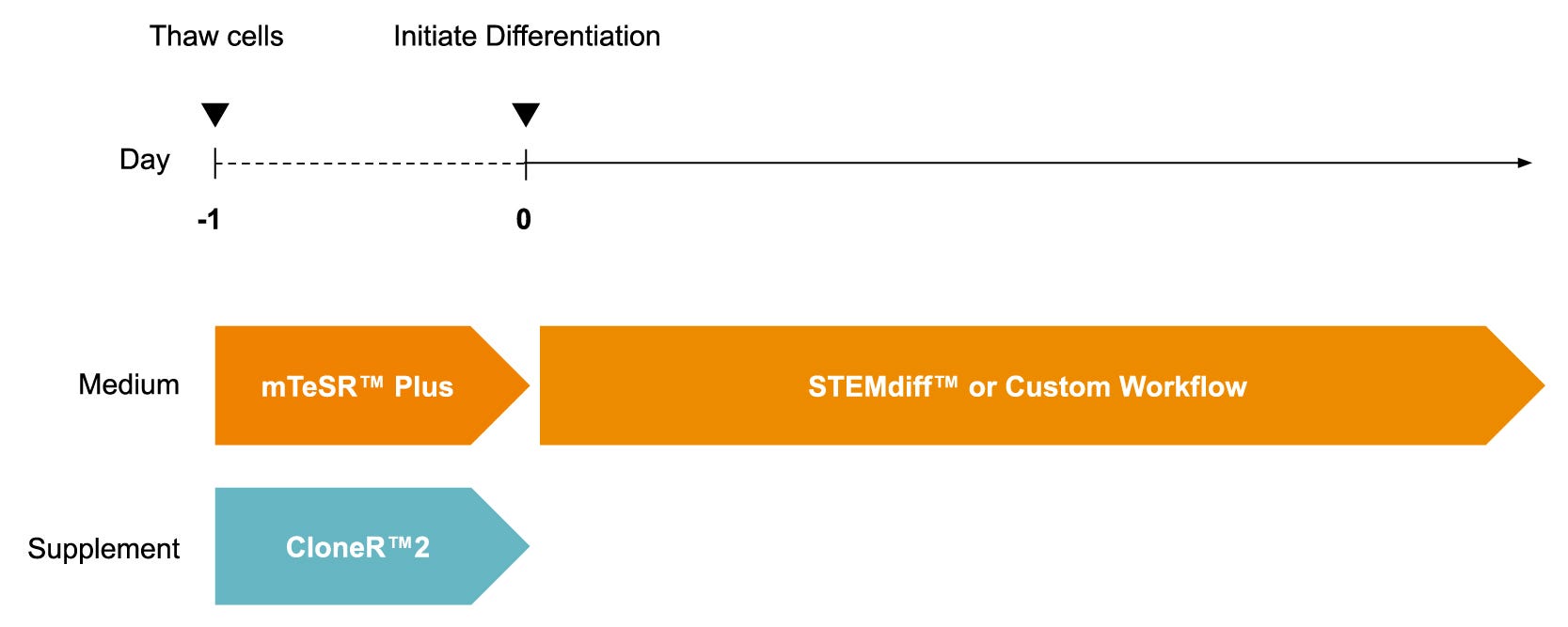
Figure 1. Schematic for a Generalized Monolayer Protocol to Thaw and Culture iPSCdirect™ for iPSC-Based Monolayer Workflows
iPSCdirect™ cells can be thawed and plated into mTeSR™ Plus (Catalog #100-0276) supplemented with CloneR™2 (Catalog #100-0691) and incubated overnight according to product instructions. Recommendations for seeding densities to reach the desired confluency at 24 hours may be found in the Product Information Sheet. After 24 hours, cells are ready for STEMdiff™ or customized monolayer workflows. iPSC = induced pluripotent stem cell.
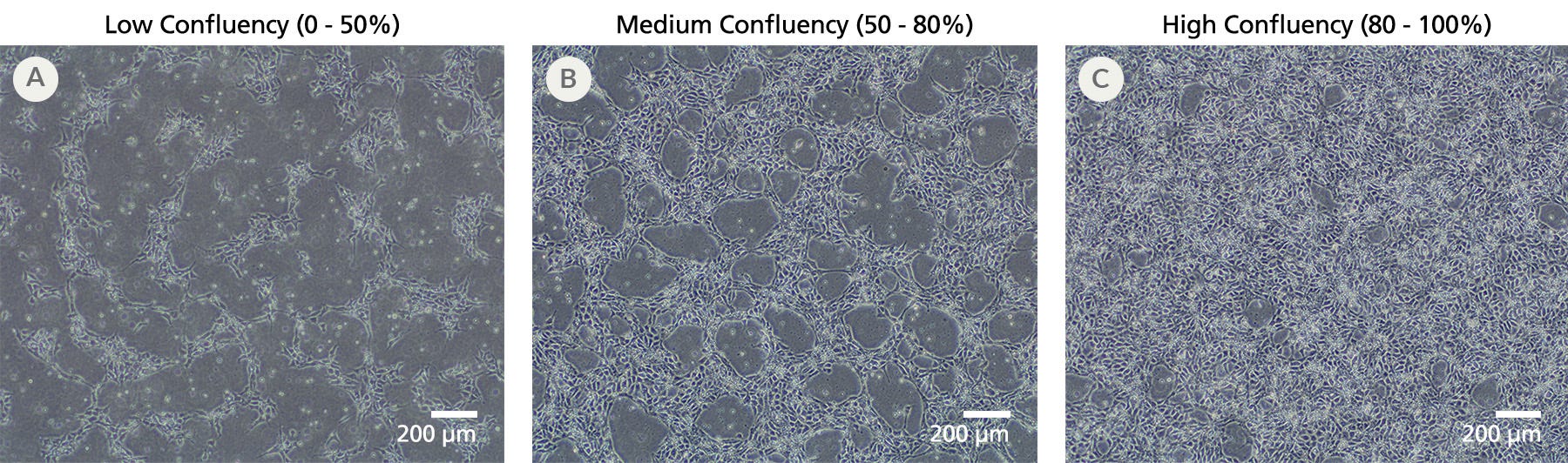
Figure 2. iPSCdirect™ SCTi003-A Cells Can Be Seeded to Reach a Range of Confluencies After 24 Hours
To reach the desired confluency for downstream experiments, thaw and plate iPSCdirect™ cells into mTeSR™ Plus with CloneR™2 (Catalog #100-0276 and #100-0691) at the densities recommended in the Product Information Sheet. These representative examples of (A) low confluency, (B) medium confluency, and (C) high confluency were cultured after thaw on Corning® Matrigel® hESC-Qualified Matrix and imaged at a magnification of 4X.
Table 1. iPSC Line SCTi003-A Is Derived from a Healthy Female Donor
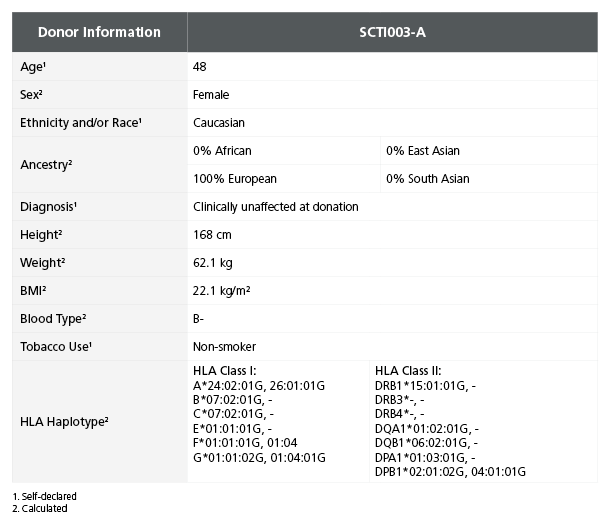
Demographic, health, and genetic characteristics of the SCTi003-A donor are compiled based on self-reported information and whole-exome sequencing. Sex was determined by karyotype. Ancestry and HLA haplotype were calculated from whole-genome and whole-exome sequencing combined data. Blood type (ABO/Rh blood group) was determined by next-generation sequencing. Height, weight, and BMI were calculated at the donation facility. iPSC = induced pluripotent stem cell.
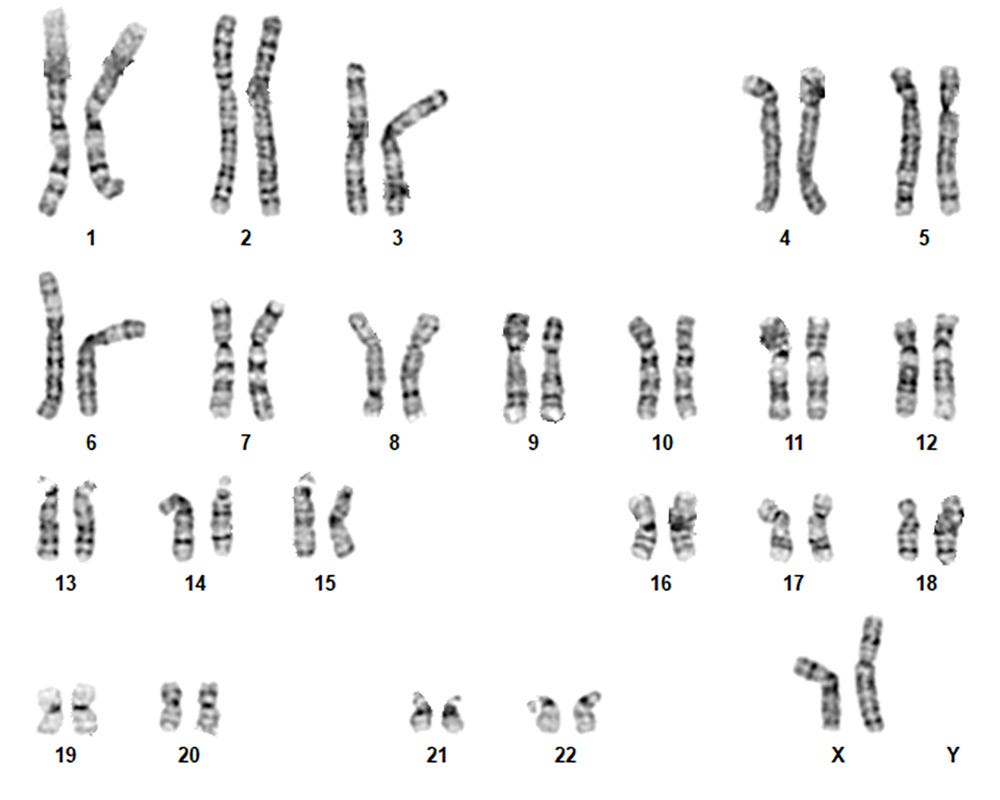
Figure 3. iPSCdirect™ SCTi003-A Human Pluripotent Stem Cells Maintain a Typical Karyotype
G-T-L banding for thawed iPSCdirect™ cells (SCTi003-A p34, n = 40) shows a typical karyotype with no evidence of clonal abnormalities at a band resolution of 425 - 475 G-bands per haploid genome.
Table 2. Single Nucleotide Polymorphism Microarray Analysis Characterizes SCTi003-A Copy Number Variants
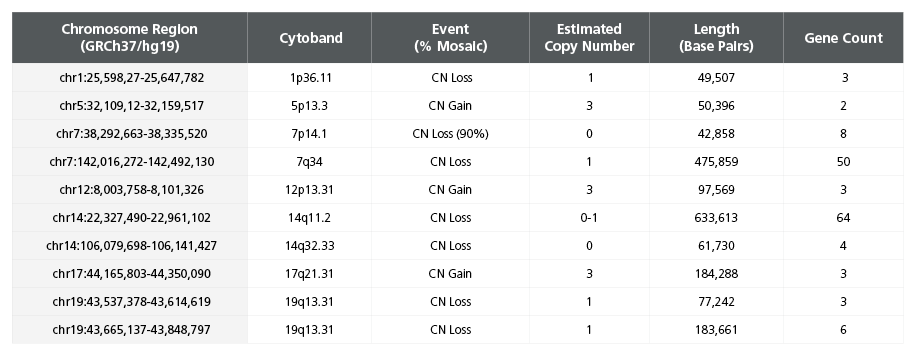
DNA was extracted from a vial of SCTi003-A from the Master Cell Bank and subject to SNP microarray analysis to identify large-scale copy number variants (CNVs). The cells display two reportable CNVs, defined as those greater than 400kb in size, on chromosome 7 and 14 (rows highlighted in bold font). These losses are located in the TCR regions of the genome and are indicative of VDJ recombination process during T Cell development. Array design, genomics position, genes, and chromosome banding are based on genome build GRCh37/hg19. chr = chromosome; start cyto = cytogenetic band at the start of the base pair imbalance; end cyto = cytogenetic band at the end of the base pair imbalance; bp = base pairs; SNP = single nucleotide polymorphism; TCR = T Cell Receptor; VDJ = variable, diversity, joining segment.
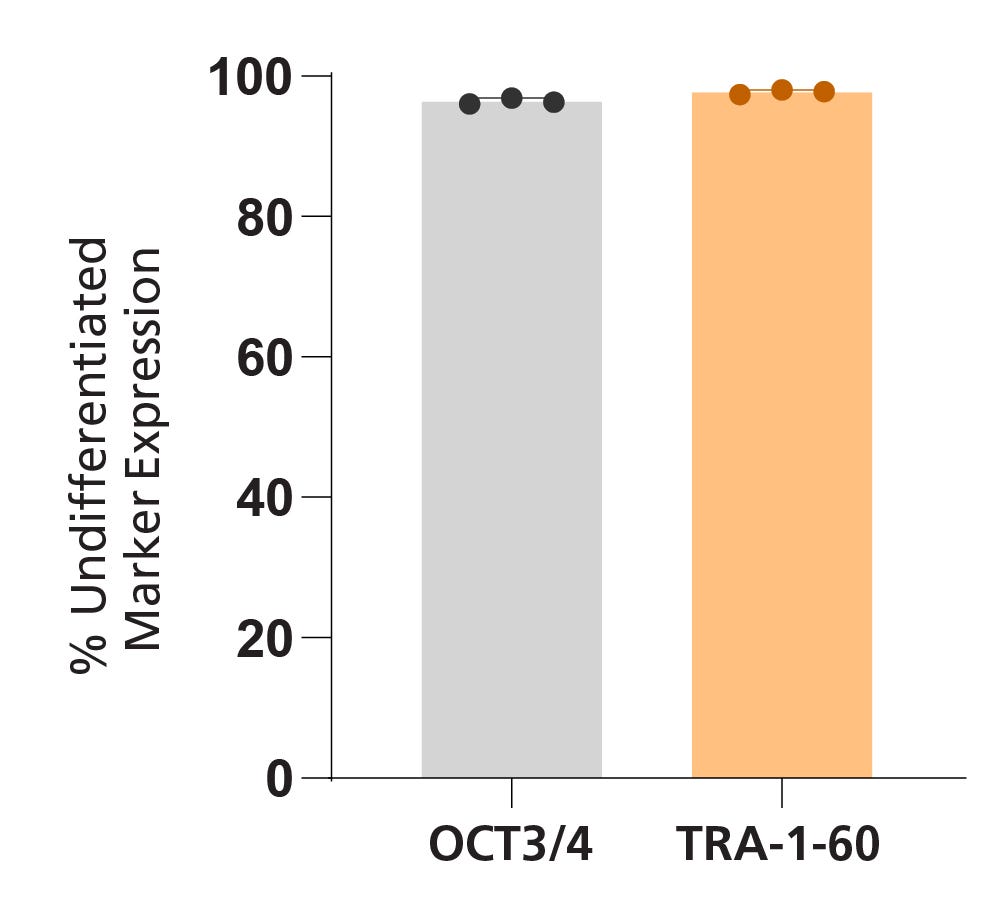
Figure 4. iPSCdirect™ SCTi003-A Cells Express Undifferentiated Cell Markers
iPSCdirect™ SCTi003-A was characterized using flow cytometry for undifferentiated cell markers OCT3/4 and TRA-1-60. Percentage marker expression was quantified 4 days after thawing, from analyses of three biological replicates (n = 3 vials). Error bars represent the standard deviation.
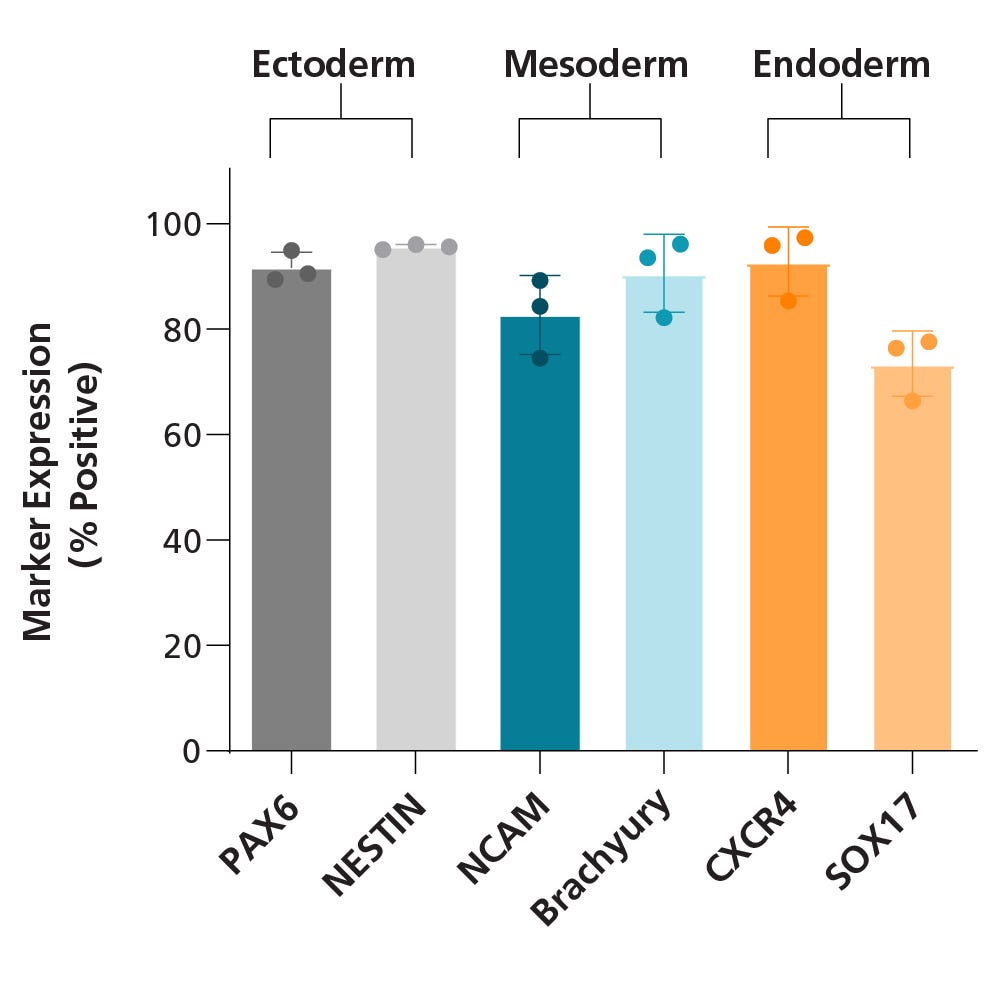
Figure 5. iPSCdirect™ SCTi003-A Human Pluripotent Stem Cells Demonstrate a High Trilineage Differentiation Capacity
iPSCdirect™ SCTi003-A cells were split into 3 groups, differentiated using STEMdiff™ Trilineage Differentiation Kit (Catalog #05230), and then subjected to flow cytometry analysis. Two markers for each embryonic germ layer were assessed, and bars represent mean marker expression for each group of cells (dots represent the average of 3 technical replicates; error bars represent standard deviation; n = 3 biological replicates). All lineage-specific markers were expressed by more than 70% of differentiated cells. Expression of PAX6 and Nestin confirm differentiation to the ectoderm lineage, NCAM and Brachyury (T) expression confirm differentiation to the mesoderm lineage, and CXCR4 and SOX17 expression confirm differentiation to the endoderm lineage.
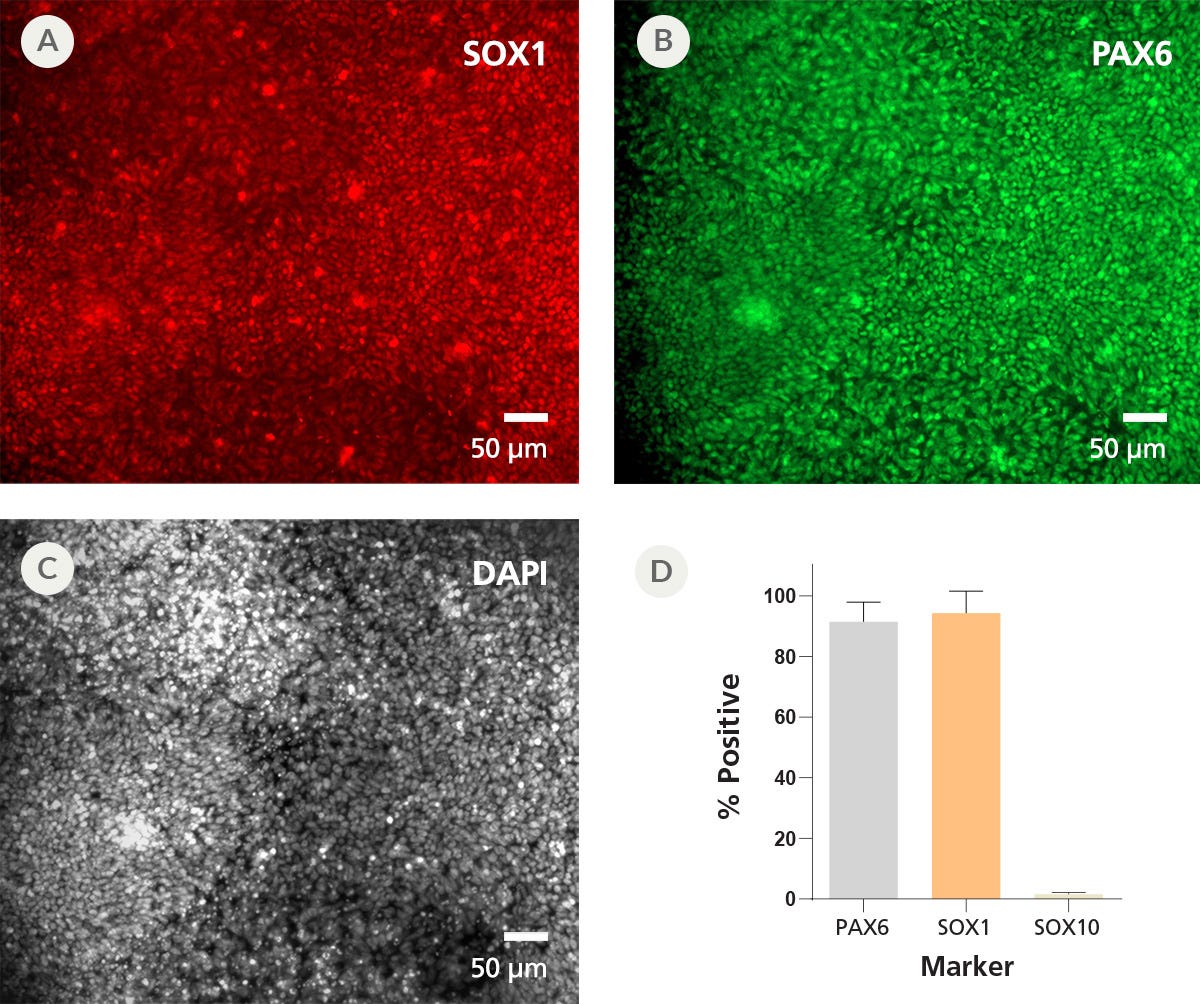
Figure 6. iPSCdirect™ SCTi003-A Human Pluripotent Stem Cells Can Efficiently Differentiate into Neural Progenitor Cells Upon Thawing
NPCs were generated from iPSCdirect™ SCTi003-A cells using the protocol in the iPSCdirect™ Product Information Sheet followed by the monolayer protocol from Day 1 in the STEMdiff™ SMADi Neural Induction Kit Technical Manual (Catalog #08581). After the first 7 days of the protocol, the resulting NPCs were fixed for immunocytochemistry. The NPCs expressed neural progenitor markers (A) SOX1 and (B) PAX6, and nuclei were visualized with (C) DAPI. (D) Marker expression was quantified and negative control SOX10 expression was minimal. Error bars represent standard deviation (n = 2 biological replicates). NPCs = neural progenitor cells.
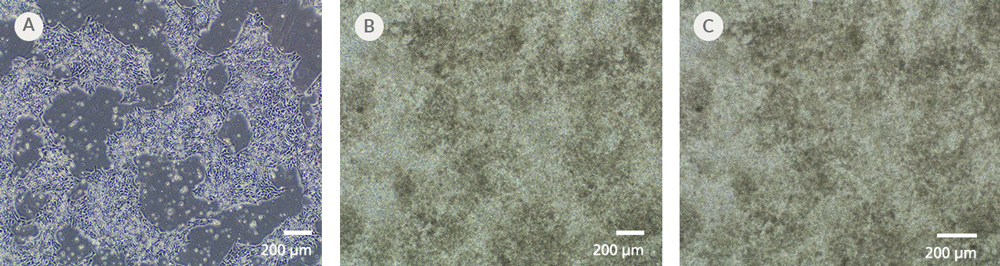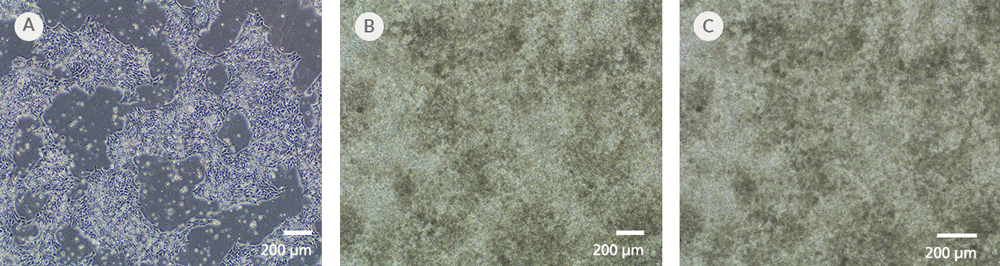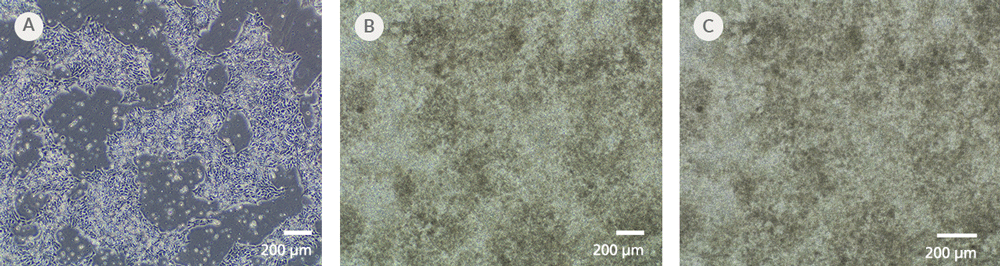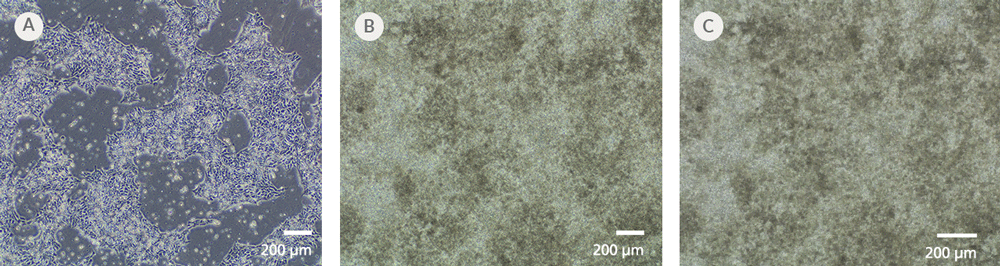
Figure 7. iPSCdirect™ SCTi003-A Human Pluripotent Stem Cells Can Successfully Differentiate into Ventricular Cardiomyocytes
Ventricular cardiomyocytes were generated from iPSCdirect™ SCTi003-A cells using STEMdiff™ Ventricular Cardiomyocyte Differentiation Kit (Catalog #05010). (A) 48 hours after thawing and plating in mTeSR™ Plus and CloneR™2, iPSCdirect™ cells reached the desired confluency and are ready for Day 0 of differentiation according to the STEMdiff™ Ventricular Cardiomyocyte Product Information Sheet. (B) By Day 15 of differentiation, monolayer cultures show iPSC-derived ventricular cardiomyocytes that (C) exhibit coordinated beating behavior.
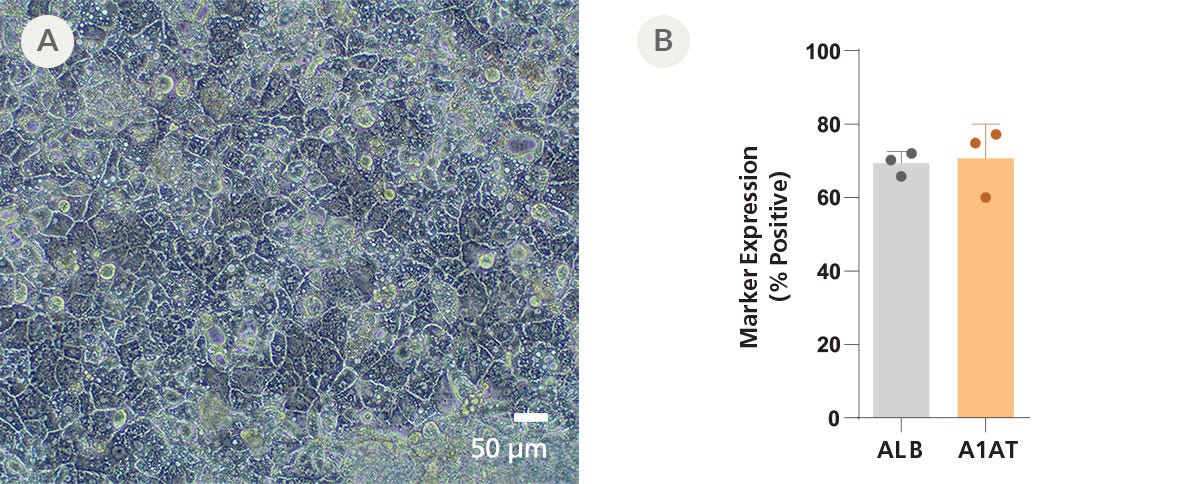
Figure 8. iPSCdirect™ SCTi003-A Human Pluripotent Stem Cells Can Successfully Differentiate into Hepatocytes
iPSCdirect™ SCTi003-A cells were plated according to the iPSCdirect™ Product Information Sheet, in preparation for Day 1 of the STEMdiff™ Hepatocyte Kit (Catalog #100-0520) protocol outlined in the STEMdiff™ Hepatocyte Kit Product Information Sheet. (A) Monolayers of hepatocyte-like cells with characteristic polygonal morphology and binucleation are apparent by Day 21 of differentiation. (B) Marker expression was quantified by flow cytometry for hepatic markers albumin (ALB) and alpha1-antitrypsin (A1AT). Error bars represent standard deviation (n = 3 biological replicates).
Find supporting information and directions for use in the Product Information Sheet or explore additional protocols below.
This product is designed for use in the following research area(s) as part of the highlighted workflow stage(s). Explore these workflows to learn more about the other products we offer to support each research area.
Thank you for your interest in IntestiCult™ Organoid Growth Medium (Human). Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration
| Species | Human |
|---|---|
| Contains | FreSR™-S |
| Cell And Tissue Source | Pluripotent Stem Cells |
| Donor Status | Normal |

cGMP,稳定的人类胚胎干细胞和iPS细胞维持培养基

为提高人类胚胎干细胞和iPS细胞在单细胞工作流程中的存活率而定义的补充

人多能干细胞系,冷冻

与 TeSR™ 维持培养基配合使用,用于维持人胚胎干细胞(ES)和诱导多能干细胞(iPS)的基质
扫描二维码或搜索微信号STEMCELLTech,即可关注我们的微信平台,第一时间接收丰富的技术资源和最新的活动信息。
如您有任何问题,欢迎发消息给STEMCELLTech微信公众平台,或与我们通过电话/邮件联系:400 885 9050 INFO.CN@STEMCELL.COM。

